Welcome to BioNeuralNet Beta 0.1
Note: This is a beta version of BioNeuralNet. It is under active development, and certain features may be incomplete or subject to change. Feedback and bug reports are highly encouraged to help us improve the tool.
BioNeuralNet is a Python-based software tool designed to streamline the transformation of multi-omics data into network-based representations and lower-dimensional embeddings, enabling advanced analytical processes like clustering, feature selection, disease prediction, and environmental exposure assessment. Python Installation via pip:
pip install bioneuralnet==0.1.0b1
For installation details and other options, go to Installation.
Example: Transforming Multi-Omics for Enhanced Disease Prediction
View full-size image: Transforming Multi-Omics for Enhanced Disease Prediction
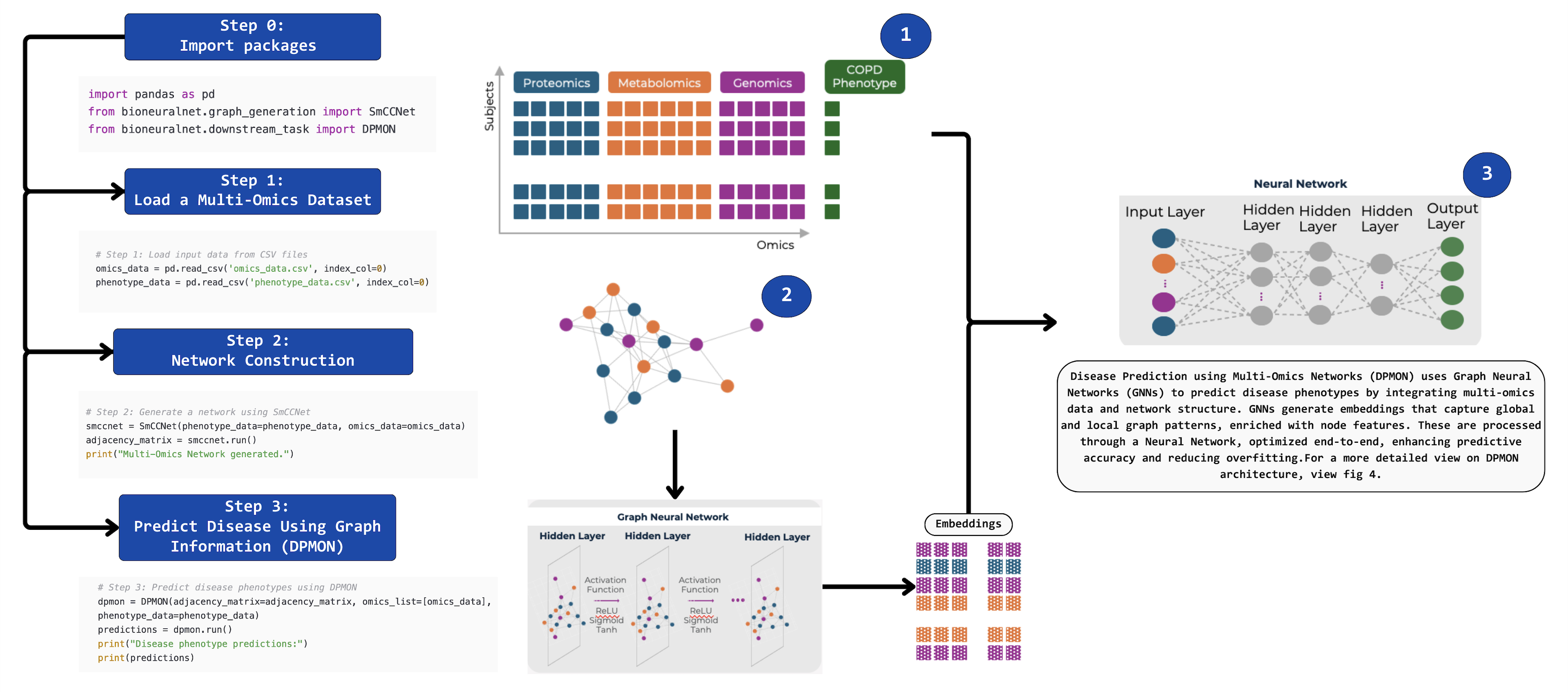
BioNeuralNet: Transforming Multi-Omics for Enhanced Disease Prediction
BioNeuralNet enables seamless integration of multi-omics data into a network-based analysis pipeline. Here is a quick example demonstrating how to generate a network representation using SmCCNet and apply it to disease prediction using DPMON:
Note:
Data Preparation:
Input your multi-omics data (e.g., proteomics, metabolomics, genomics) along with phenotype data.
Network Construction:
Use Sparse Multiple Canonical Correlation Network (SmCCNet) to generate a network from the omics data.
This step constructs an adjacency matrix capturing correlations and interactions between features.
Disease Prediction:
Disease Prediction using Multi-Omics Networks (DPMON) uses Graph Neural Networks (GNNs) to predict disease phenotypes.
Integrates multi-omics data and network structure information to generate GNNs embeddings that capture global and local graph patterns.
It enhances the Omics-data by creating enriched with node features. These are processed through a Neural Network, optimized end-to-end, enhancing predictive accuracy and reducing overfitting.
Code Example:
import pandas as pd
from bioneuralnet.graph_generation import SmCCNet
from bioneuralnet.downstream_task import DPMON
# Step 1: Load Multi-Omics Dataset
omics_data = pd.read_csv('omics_data.csv', index_col=0)
phenotype_data = pd.read_csv('phenotype_data.csv', index_col=0)
# Step 2: Generate a network using SmCCNet
smccnet = SmCCNet(phenotype_data=phenotype_data, omics_data=omics_data)
adjacency_matrix = smccnet.run()
print("Multi-Omics Network generated.")
# Step 3: Enhanced disease prediction using network information with DPMON
dpmon = DPMON(adjacency_matrix=adjacency_matrix, omics_list=[omics_data], phenotype_data=phenotype_data)
predictions = dpmon.run()
print("Disease phenotype predictions:")
print(predictions)
Output:
Adjacency Matrix: The network representation of the multi-omics data.
Predictions: Disease phenotype predictions for each sample.
BioNeuralNet Overview
Looking to explore the capabilities of BioNeuralNet? Here is a brief overview of the key components:
Graph Construction: Build multi-omics networks using methods like Weighted Gene Co-expression Network Analysis (WGCNA), Sparse Multiple Canonical Correlation Network (SmCCNet), or import existing networks.
Graph Clustering: Identify functional modules and communities using hierarchical clustering, PageRank, or Louvain clustering algorithms.
Network Embedding: Generate embeddings with Graph Neural Networks (GNNs) or Node2Vec, simplifying high-dimensional data into lower-dimensional representations.
Subject Representation: Integrate embeddings into omics data to enrich subject-level features, enhancing the dataset for downstream analyses.
Downstream Tasks: Perform advanced analyses like disease prediction using network information. Seamlessly integrate your own downstream tasks by leveraging existing components.
View full-size image: BioNeuralNet Overview
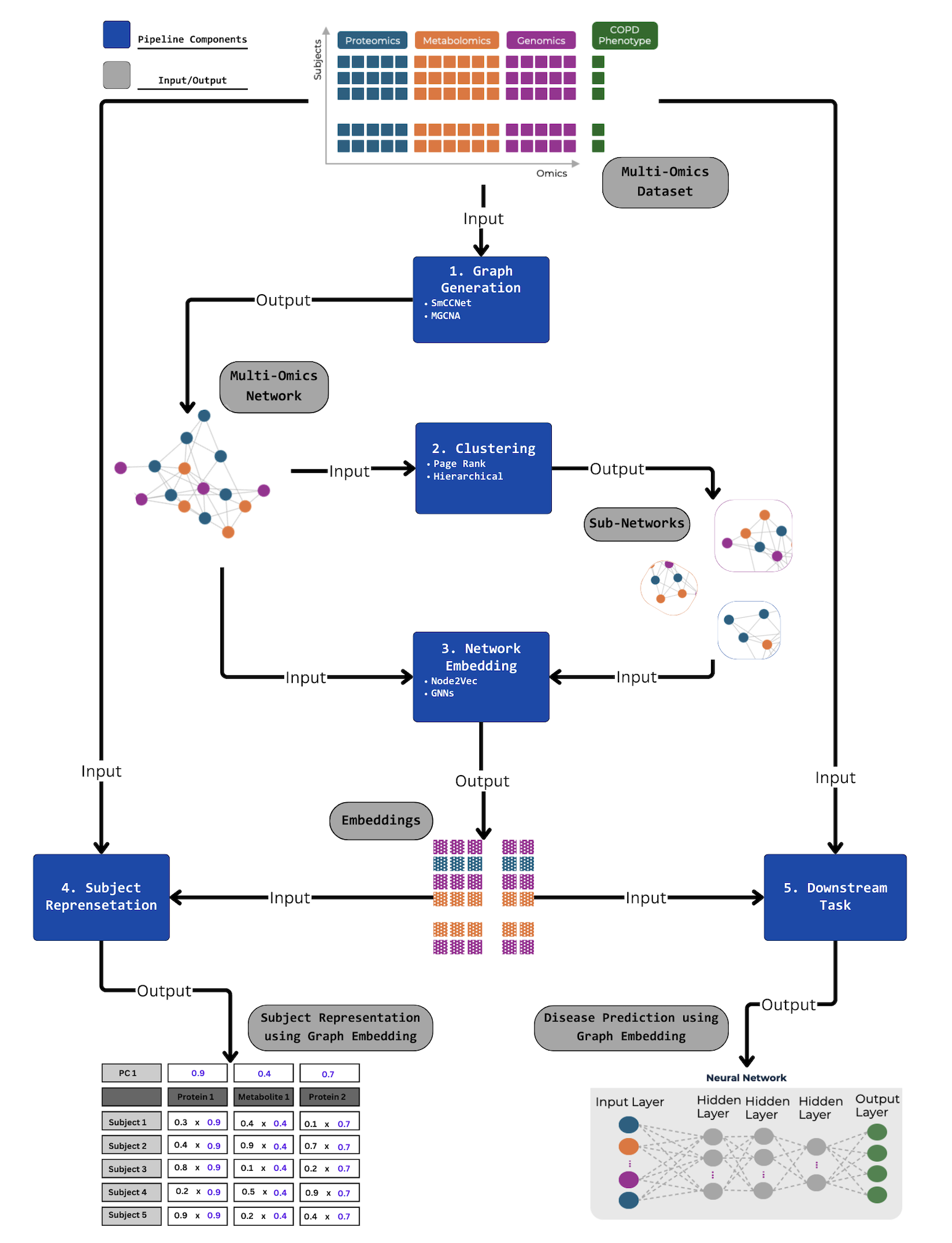
BioNeuralNet Overview
Subject Representation
View full-size image: Subject Representation
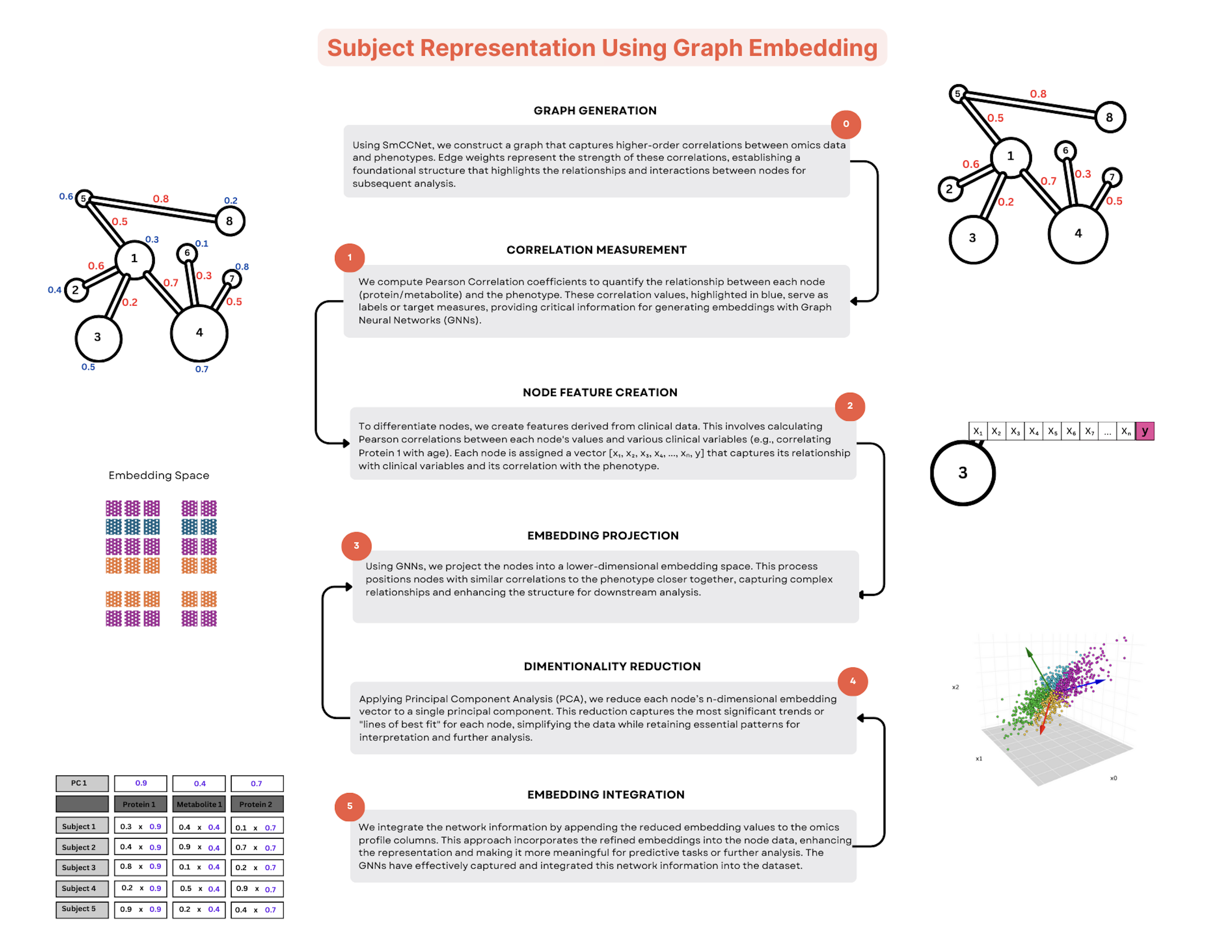
Subject-level embeddings provide richer phenotypic and clinical context.
Disease Prediction
View full-size image: Disease Prediction (DPMON)
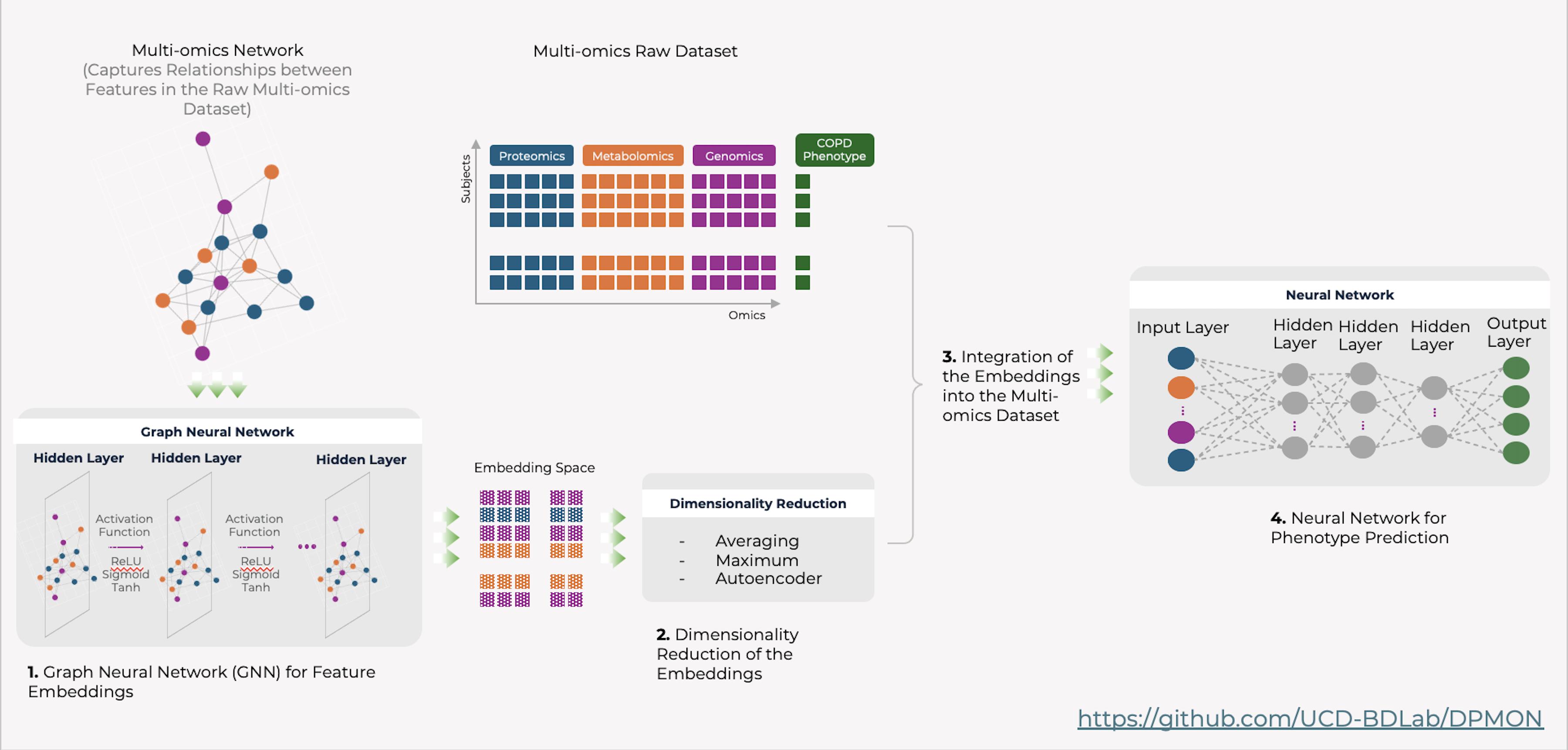
Embedding-enhanced subject data using DPMON for improved disease prediction.
Documentation Overview
Contents:
- Installation
- Tutorials
- Example 1: Sparse Multiple Canonical Correlation Network (SmCCNet) Workflow with Graph Neural Network (GNN) Embeddings
- Example 2: Weighted Gene Co-expression Network Analysis (WGCNA) Workflow with Graph Neural Network (GNN) Embeddings
- Example 3: Disease Prediction Using Graph Information (SmCCNet + DPMON)
- Example 4: Sparse Multiple Canonical Correlation Network (SmCCNet) + PageRank Clustering + Visualization
- Tools
- User API
- Frequently Asked Questions (FAQ)